How to Build a Custom Estimator for scikit-learn
This post will show you how to implement your own model and make it compliant with scikit-learn’s API. The final result will be a model that can not only be fitted and used for predictions but also be used in combination with other scikit-learn tools like grid search and pipelines.
Introduction
This post traces back to a few months ago. In one of my machine learning courses, we were discussing the topic of imbalanced data and how algorithms have a hard time learning when data is not balanced. For our learning algorithms, the event we are trying to predict is so uncommon that in the process of improving evaluation scores, our model will rarely predict this category. As soon as I came into contact with this topic I knew it was super important. I had seen imbalanced data before and knew very well that real-life is not as easy as toy datasets. The case that resonated the most with me was predicting customer churn.
Anyway, in our class, we saw many ways we could avoid or try to mitigate the problem of imbalanced data, but one solution was particularly interesting to me: Ensemble Resampling. We were following Andreas Müller’s lectures. In this section, he mentions a paper “Exploratory Undersampling for Class-Imbalance Learning”. In this paper, the authors describe an interesting approach to improving classification in imbalanced datasets, they call it EasyEnsemble. The basic idea is to train an ensemble model using under-sampling, resampling every time, and combine the overall result of the ensemble. The benefit of this model is that it reduces the amount of data that gets discarded in the process of under-sampling.
Building the Model
The specific implementation of your custom model will depend a lot on the model you are trying to extend or build upon. If you are starting from scratch, a good start is the BaseEstimator. We can start by building the class for our model:
class ResampledEnsemble(BaseEstimator):
def __init__(self):
passThere are a few methods we need to implement, but first and foremost, we can go ahead and create that __init__ method.
def __init__(self, base_estimator=DecisionTreeClassifier(), n_estimators=100,
max_depth=None, max_features=None, min_samples_split=2, min_samples_leaf=1):
self._estimator_type = "classifier"
self.base_estimator = base_estimator
self.n_estimators = n_estimators
self.max_depth = max_depth
self.max_features = max_features
self.min_samples_split = min_samples_split
self.min_samples_leaf = min_samples_leafAt this point, we are basically just passing on all the arguments and storing them as attributes. There is a couple of other things we need to do to implement the actual model. We need a way to generate the ensemble. To accomplish this, we’ll define a function to generate the estimators: _generate_estimators.
def _generate_estimators(self):
estimators = []
for i in range(self.n_estimators):
est = clone(self.base_estimator)
est.random_state = i
est.max_depth = self.max_depth
est.max_features = self.max_features
est.min_samples_split = self.min_samples_split
est.min_samples_leaf = self.min_samples_leaf
pipe = make_imb_pipeline(
RandomUnderSampler(random_state=i, replacement=True),
est
)
estimators.append((f"est_{i}", pipe))
return estimatorsThis method does not take any argument because all the data we need is already stored in the instance’s attributes. We start by creating a list of estimators. This will be our end result; a list of estimators. The number of estimators is determined by the attribute self.n_estimators, so we just create each estimator inside a for-loop. To build each estimator, we clone the self.base_estimator, set it up using all the parameters and make a pipeline with it. Instead of using scikit-learn’s pipeline, we need to use imbalanced-learn’s pipeline (docs) so that we can do our under-sampling.
Now we can go back to the __init__ function and generate these estimators:
def __init__(
self,
base_estimator=DecisionTreeClassifier(),
n_estimators=100,
max_depth=None,
max_features=None,
min_samples_split=2,
min_samples_leaf=1,
):
self._estimator_type = "classifier"
self.base_estimator = base_estimator
self.n_estimators = n_estimators
self.max_depth = max_depth
self.max_features = max_features
self.min_samples_split = min_samples_split
self.min_samples_leaf = min_samples_leaf
self.estimators = self._generate_estimators()Perfect, now the last piece of the puzzle is to generate a single estimation that makes a decision about the result from the ensemble. We’ll use the VotingClassifier that will take the result of the estimators and reach a final conclusion using soft voting.
def __init__(
self,
base_estimator=DecisionTreeClassifier(),
n_estimators=100,
max_depth=None,
max_features=None,
min_samples_split=2,
min_samples_leaf=1,
):
self._estimator_type = "classifier"
self.base_estimator = base_estimator
self.n_estimators = n_estimators
self.max_depth = max_depth
self.max_features = max_features
self.min_samples_split = min_samples_split
self.min_samples_leaf = min_samples_leaf
self.estimators = self._generate_estimators()
self.estimator = VotingClassifier(self.estimators, voting="soft")Finally, there are a few other methods we need to implement for this to work but don’t worry, that was the hard part the rest is fairly straightforward.
We’ll implement fit, predict and classes_ methods that we’ll simply delegate responsibility by calling the VotingClassifier methods.
def fit(self, X, y, sample_weight=None):
return self.estimator.fit(X, y, sample_weight)
def predict(self, X):
return self.estimator.predict(X)
def classes_(self):
if self.estimator:
return self.estimator.classes_Last but not least, we need to implement a set_params method. As mentioned in the docs, this functions is quite fundamental as it is used during grid searches to update the parameters of the models. In our case, this is fairly straightforward:
def set_params(self, **params):
if not params:
return self
for key, value in params.items():
if hasattr(self, key):
setattr(self, key, value)
else:
self.kwargs[key] = value
self.estimators = self._generate_estimators()
self.estimator = VotingClassifier(self.estimators, voting="soft")
return selfPutting it all together, we get our own model that we can use in combination with other scikit models and tools. We can use it in a pipeline, grid search and score it like any other model.
The only thing left to do is test it out!
Using the Model
If you store the class we just built in a resampled_ensemble.py file, importing your model is just as simple as:
from resampled_ensemble import ResampledEnsembleFrom now on, it’s basically just like a scikit-learn model, so we can proceed the usual way:
data = load_breast_cancer(as_frame=True)
X_train, X_test, y_train, y_test = train_test_split(data.data, data.train, random_state=0)We’ll instantiate the model, fit it and check how it performed:
re = ResampledEnsemble()
re.fit(X_train, y_train)
y_pred = re.predict(X_test)
classification_report(y_test, y_pred)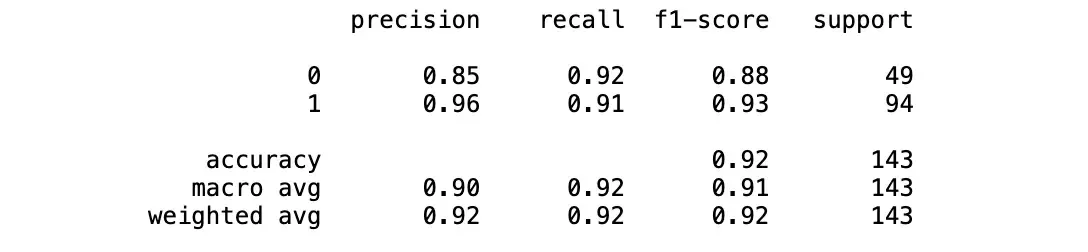
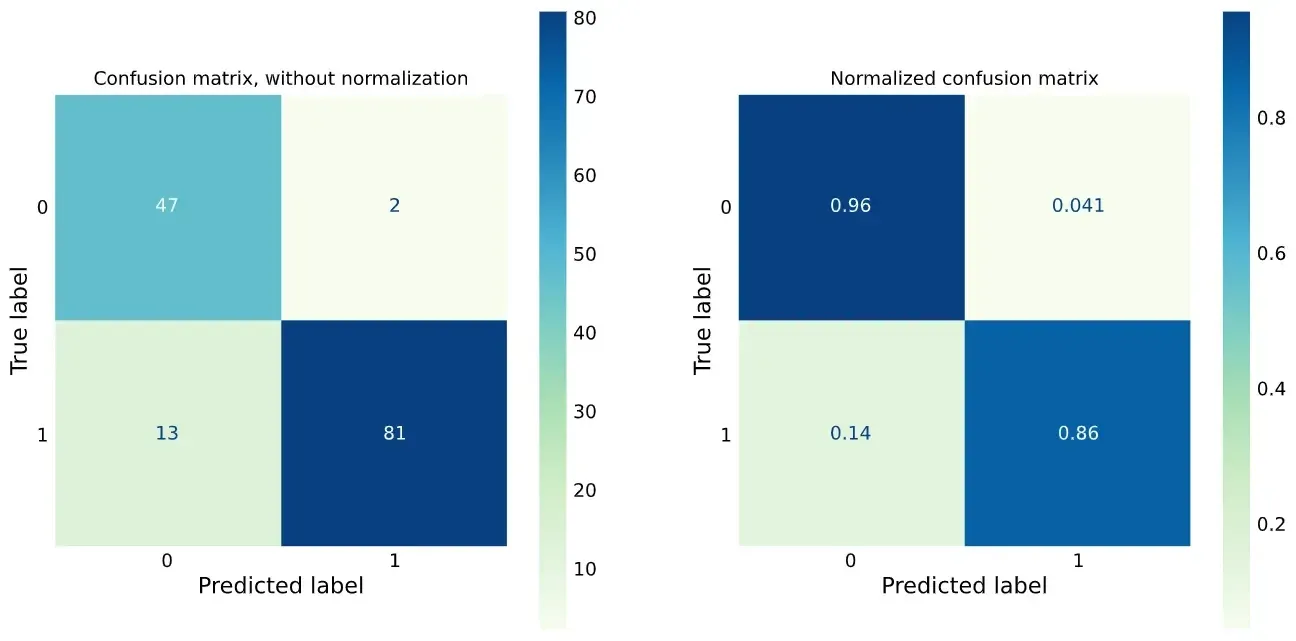
We can also plot the ROC curve and confusion matrices:
plot_roc_curve(re, X_test, y_test, ax=ax);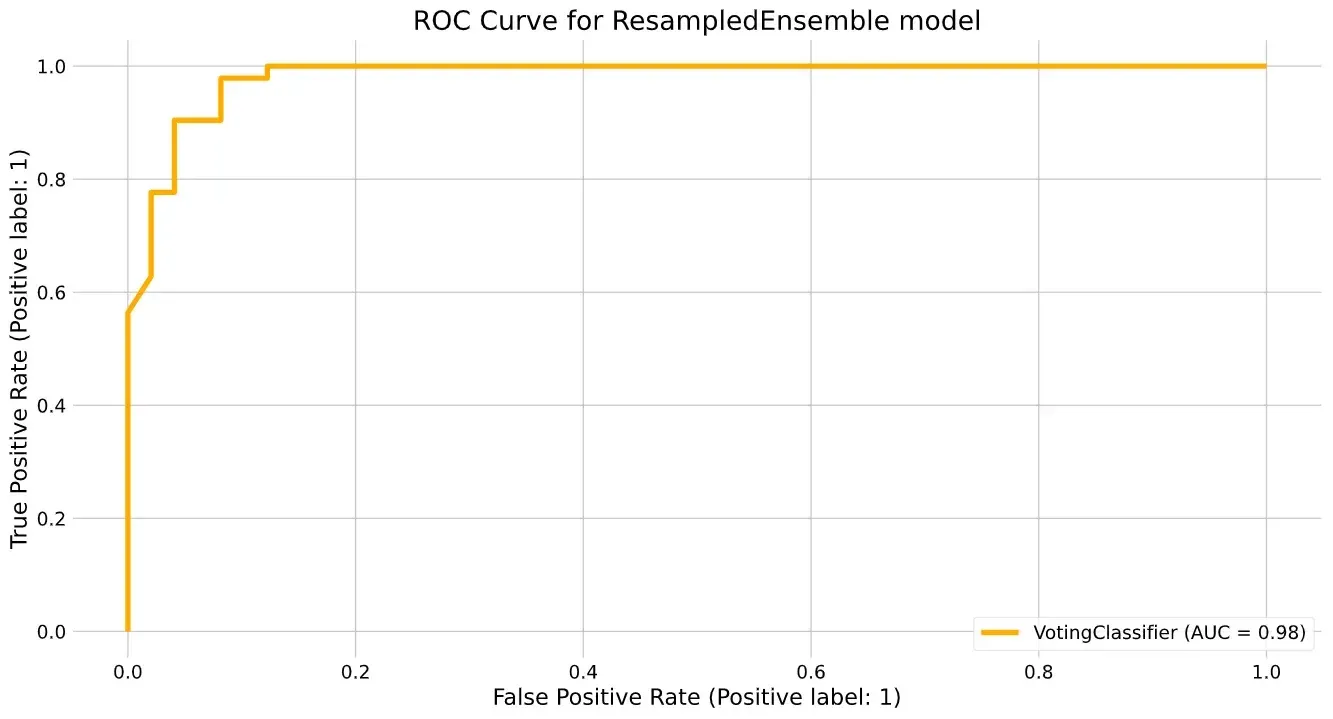
plot_confusion_matrix(
re,
X_test,
y_test,
display_labels=[0, 1, 2],
cmap=plt.cm.GnBu,
normalize=None,
ax=ax1,
)
plot_conf = plot_confusion_matrix(
re,
X_test,
y_test,
display_labels=[0, 1, 2],
cmap=plt.cm.GnBu,
normalize="true",
ax=ax2,
)
However, I think the best part is that we can now use it in pipelines and grid searches:
pipe = make_pipeline(
SimpleImputer(missing_values=np.nan, strategy="mean"),
MinMaxScaler(),
ResampledEnsemble(
max_features="auto",
min_samples_split=0.01,
min_samples_leaf=0.0001,
n_estimators=300,
),
)
grid_params = {
"resampledensemble__max_depth": np.linspace(5, 40, 3, endpoint=True, dtype=int),
}
grid = GridSearchCV(
pipe, grid_params, cv=4, return_train_score=True, n_jobs=-1, scoring="f1_macro"
)
grid.fit(X_train, y_train)
best_score = grid.best_score_
best_params = grid.best_params_Conclusion
As you can see, it’s extremely simple to build your own custom models for scikit-learn while also taking advantage of the other tools scikit-learn has to offer. You can build whichever model you want and still be able to use it with metrics, pipelines, and grid searches. In this post, I showed you one example of implementing a model for imbalanced data, but the possibilities are truly endless. For example, this same methodology could be used to implement Bayesian models of probabilistic programming in scikit-learn. I guess we’ll have to discuss it in a future post…
Liu, X. Y., Wu, J., & Zhou, Z. H. (2008). Exploratory undersampling for class-imbalance learning. IEEE Transactions on Systems, Man, and Cybernetics, Part B (Cybernetics), 39(2), 539-550.
Müller, A. (2018). Advanced Scikit-learn, GitHub repository, https://github.com/amueller/ml-training-advanced
Müller, A. C., & Guido, S. (2016). Introduction to Machine Learning with Python: A Guide for Data Scientists (1st ed.). O’Reilly Media.